Biophysical Chemistry, Kyoto Prefectural University
Molecular Recognition Mechanisms by Proteins
-Protein Structure, Flexibility, and Function-
Masayuki Oda, Ph.D.
Graduate School of Life and Environmental Sciences, Kyoto Prefectural University
1-5, Hangi-cho, Shimogamo, Sakyo-ku, Kyoto 606-8522, Japan
TEL&FAX: +81-75-703-5673, E-mail: oda@kpu.ac.jp
【Researchmap】 【ORCiD】
1. Members;

Masayuki Oda, Ph.D. (Professor)
Fumiya Kondo (M2)
Saaya Yabuno (M2)
Mutsumi Yoshida (M2)
Haruka Kawase (M1)
Shinya Nishatani (M1)
Mei Suzuki (B4)
Hitomi Nishida (B4)
Yui Maruta (B4)
Qinghua Wang (Research Student)
Kosuke Morikawa, Ph.D.
(Senior Researcher)
Masayuki Takahashi, Ph.D.
(Senior Researcher)
Yoshiji Hantani, Ph.D. (Senior Researcher)
Daniela Battaglia Hirata, Ph.D. (Senior Researcher)
Alumni;
Saki Ochi, Yumi Kitagawa, Ayako Shiota, Rento Uegaki, Riri Takenaka, Momoka Iiyama, Miho Emori, Takahiro Hayashi, Tomoyo Hira, Iori Murata, Maki Kawasaki, Akihiro Nishiguchi, Zengwei Liao, Masahiro Tanaka, Ikuko Iizumi, Akane Senga, Yuhi Hosoe, Takanori Yamaoka, Toshifumi Fujioka, Michiko Oka, Yuri Yamagami, Satomi Inaba Ph.D., Daiki Usui, Ayako Miki, Yusui Sato, Shohey Shimizu, Tomonari Tamashiro, Nobutaka Komichi, Yoko Shima, Shoko Motoki, Koki Yamamoto, Kazuki Hayashida, Chika Higashiura, Yoichi Tanabe Ph.D, Yusuke Tanaka, Tomoki Sano, Tomoyuki Sunahashi, Kotaro Hara, Asami Maruoka, Yuichiro Takagi, Kuniomi Nakamura, Yasutomo Inui, Hiromi Asada, Hidekazu Kaminaka, Keiko Watanabe
2. Research;
 My research interests are related to understanding the correlation between protein structure and function. We specifically focus on the protein flexibility, which is difficult to determine but is critical for understanding protein structure-function relationship.
My research interests are related to understanding the correlation between protein structure and function. We specifically focus on the protein flexibility, which is difficult to determine but is critical for understanding protein structure-function relationship.
To achieve our projects, we purify proteins, and analyze the physical properties and molecular interactions using biophysical methods such as circular dichroism (CD), isothermal titration calorimetry (ITC), and surface plasmon resonance (SPR) biosensor.
We analyze 3D structures and structural dynamics at atomic resolution using X-ray crystallography and nuclear magnetic resonance (NMR). We also use dynamic light scattering (DLS), differential scanning calorimetry (DSC), electron microscopy (EM), small-angle X-ray scattering (SAXS), and diffracted X-ray tracking (DXT) for analyses.
The results can be applied in protein engineering to increase protein stability and function, and further, for rational design of small molecules to inhibit molecular interactions. The present targets for protein interactions are as follows.
2-1. Antibody
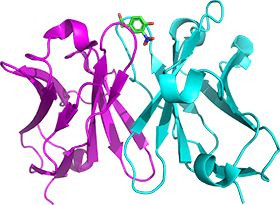 Antibodies can recognize various shapes and sizes of antigens and have been used as drugs. We overexpress and purify single-chain Fv (scFv) antibodies, and analyze the structural dynamics and the antigen recognition mechanism. We also focus on the affinity maturation of antibody. We generate mutants of scFv to analyze the role of residue to increase the antigen-binding affinity.
Antibodies can recognize various shapes and sizes of antigens and have been used as drugs. We overexpress and purify single-chain Fv (scFv) antibodies, and analyze the structural dynamics and the antigen recognition mechanism. We also focus on the affinity maturation of antibody. We generate mutants of scFv to analyze the role of residue to increase the antigen-binding affinity.
2-2. CD28 family molecules and adapter proteins
 To activate T cells, costimulatory signals via CD28-family molecules are required, together with the signals via T-cell receptors. We overexpress and purify adaptor proteins, Growth factor receptor bound protein 2 (Grb2), Grb2-related adaptor downstream of Shc (Gads), phosphatidylinositol-3 kinase (PI3K), and their SH2 domains, and analyze their molecular interactions with the intracellular regions of CD28, ICOS, and CTLA-4. The structural information can be correlated with the phenomenon of signal transduction.
To activate T cells, costimulatory signals via CD28-family molecules are required, together with the signals via T-cell receptors. We overexpress and purify adaptor proteins, Growth factor receptor bound protein 2 (Grb2), Grb2-related adaptor downstream of Shc (Gads), phosphatidylinositol-3 kinase (PI3K), and their SH2 domains, and analyze their molecular interactions with the intracellular regions of CD28, ICOS, and CTLA-4. The structural information can be correlated with the phenomenon of signal transduction.
2-3. Endo-1,3-β-glucanase
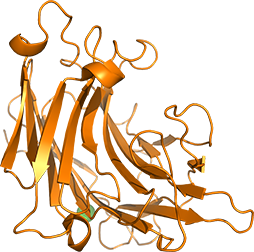 Endo-1,3-β-glucanase from Cellulosimicrobium cellulans is composed of a catalytic domain and a carbohydrate-binding module. We overexpress and purify the respective domains and their mutants, and analyze the catalytic function and the carbohydrate recognition mechanism. In addition, we also design mutants to modulate catalytic function and carbohydrate binding.
Endo-1,3-β-glucanase from Cellulosimicrobium cellulans is composed of a catalytic domain and a carbohydrate-binding module. We overexpress and purify the respective domains and their mutants, and analyze the catalytic function and the carbohydrate recognition mechanism. In addition, we also design mutants to modulate catalytic function and carbohydrate binding.
2-4. Cutinase-like enzyme (Cut190)
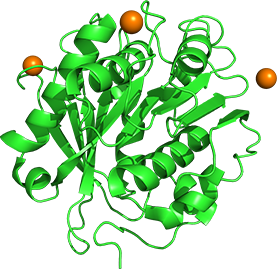 The Cut190 enzyme from Saccharomonospora viridis AHK190 can degrade the inner block of polyethylene terephthalate (PET) in the presence of Ca2+. We overexpress and purify Cut190 and its mutants to analyze the mechanism of increased activity and stability by Ca2+ binding. Crystal structural analysis so far indicates that Cut190 fluctuates in solution and the specific conformers are increased upon Ca2+ binding.
The Cut190 enzyme from Saccharomonospora viridis AHK190 can degrade the inner block of polyethylene terephthalate (PET) in the presence of Ca2+. We overexpress and purify Cut190 and its mutants to analyze the mechanism of increased activity and stability by Ca2+ binding. Crystal structural analysis so far indicates that Cut190 fluctuates in solution and the specific conformers are increased upon Ca2+ binding.
2-5. Ribonuclease HI
Escherichia coli ribonuclease HI (RNH) hydrolyzes the RNA strands of RNA/DNA hybrids in the presence of Mg2+ at the highest level, relative to other metal ions. The Mg2+ binding affinity was lower than those of other metal ions, and the low binder can express the maximum catalytic activity of RNH. The RNH activity induced upon metal ion binding closely correlates with the structural flexibility and metal ion exchange or movement.
2-6. 3α-Hydroxysteroid dehydrogenase
The NAD(P)+-dependent enzyme, 3α-HSD, catalyzes the reversible interconversion of hydroxy and oxo groups at position 3 of the steroid nucleus. We overexpress and purify 3α-HSD and its mutants, and analyze the mechanism of nucleotide-cofactor recognition and its correlation with catalytic function. We focus on the conformational change of 3α-HSD induced upon cofactor binding, and the allosteric binding of the cofactor.
2-7. DNA-binding protein
The DNA-binding domain of transcriptional factor, c-Myb, consists of three imperfect tandem repeats, R1, R2, and R3, and R2R3 is necessary for specific DNA-binding. We have shown that R2R3 fluctuates largely in solution like a semi-intrinsically disordered protein, which is critical for its function under physiological conditions. We overexpress and purify R2R3 and its mutants, and analyze the structural dynamics.
2-8. Helix-bundle protein
We design simple model peptides whose structures changes from random-coil to helix-bundle structures by forming a stable hydrophobic core in the presence of metal ions. We overexpress and purify the peptides, and analyze their structural dynamics. One of the main purposes is to correlate the structural ensemble with the information determined from single-molecule analysis.
3. Publications;
 Bekker, G.-J.*, Numoto, N.*, Kawasaki, M., Hayashi, T., Yabuno, S., Kozono, Y., Shimizu, T., Kozono, H., Ito, N., Oda, M., and Kamiya, N. (2023) Elucidation of binding mechanism, affinity and complex structure between mWT1 tumor-associated antigen peptide and HLA-A*24:02. Protein Sci. 32 (10), e4775. (*equal contributors)
Bekker, G.-J.*, Numoto, N.*, Kawasaki, M., Hayashi, T., Yabuno, S., Kozono, Y., Shimizu, T., Kozono, H., Ito, N., Oda, M., and Kamiya, N. (2023) Elucidation of binding mechanism, affinity and complex structure between mWT1 tumor-associated antigen peptide and HLA-A*24:02. Protein Sci. 32 (10), e4775. (*equal contributors)
Kitagawa, Y., Liao, Z., Morikawa, K., and Oda, M. (2023) Metal-binding and folding thermodynamics of Escherichia coli ribonuclease HI related to its catalytic function. Biophys. Chem. 295, 106961.
Ogawa, S.*, Asawa, Y.*, Iiyama, M., Yoshimori, A., Nakamura, H., and Oda, M. (2022) Regulation of CD28 binding to SH2 domains of Grb2 and PI3K by trisubstituted carboranes for T-cell activation. Bioorg. Med. Chem. Lett. 78, 129049. (*equal contributors)
Hosoe, Y.*, Miyanoiri, Y.*, Re, S.*, Ochi, S., Asahina, Y., Kawakami, T., Kuroda, M., Mizuguchi, K., and Oda, M. (2023) Structural dynamics of the N-terminal SH2 domain of PI3K in its free and CD28-bound states. FEBS J. 290 (9), 2366-2378. (*equal contributors)
Oda, M. Structural, functional, and physiological properties of anti-(4-hydroxy-3-nitrophenyl)acetyl antibodies during the course of affinity maturation. Biophys. Rev. in press.
Hayashi, T., Kamatari, Y.O., and Oda, M. (2022) Evaluation of multi-specificity of antibody G2 using its single-chain Fv and its covalently linked antigen peptides. Biophys. Chem. 290, 106893.
Iiyama, M., Hantani, Y., Wink, R.H., Kuroda, M., and Oda, M. (2022) Role of Cys residues of C-terminal SH2 domain of phosphoinositide 3-kinase in its conformational stability and CD28-binding ability. Chem. Thermodynamics Thermal Anal. 8, 100080. [PDF]
Nishiguchi, A., Murakami, A., Azuma, T., and Oda, M. (2022) A trade-off between thermostability and binding affinity of anti-(4-hydroxy-3-nitrophenyl)acetyl antibodies during the course of affinity maturation. Protein J. 41 (2), 293-303.
Ochi, S., Iiyama, M., and Oda, M. (2022) Interdomain interactions in Grb2 revealed by the conformational stability and CD28 binding analysis. Biophys. Chem. 284, 106792.
Liao, Z., Oyama, T., Kitagawa, Y., Katayanagi, K., Morikawa, K., and Oda, M. (2022) Pivotal role of a conserved histidine of Escherichia coli ribonuclease HI as proposed by X-ray crystallography. Acta Crystallogr. D78, 390-398. [PDF]
Oda, M., Sano, T., Kamatari, Y.O., Abe, Y., Ikura, T., and Ito, N. (2022) Structural analysis of hen egg lysozyme refolded after denaturation at acidic pH. Protein J. 41 (1), 71-78.
Fujioka, T.*, Numoto, N.*, Akama, H.*, Shilpa, K.*, Oka, M., Roy, P.K., Krishna, Y., Ito, N., Baker, D., Oda, M., and Tanaka, F. (2022) Varying the directionality of protein catalysts for aldol and retro-aldol reactions. ChemBioChem, 23 (2), e202100435. (*equal contributors)
Kawasaki, M. and Oda, M. (2021) Effects of chain length, temperature, and ionic strength on association and dissociation thermodynamics of DNA. Chem. Thermodyn. Thermal Anal. 3-4, 100015. [PDF]
Hosoe, Y.*, Sekiguchi, H.*, Sasaki, Y.C., and Oda, M. (2021) Structural dynamics of a DNA-binding protein analyzed using diffracted X-ray tracking. Biophys. Chem. 278, 106669. (*equal contributors)(Cover Illustration)
Tanaka, M., Kato, T., and Oda, M. (2021) Conformational changes of α-helical peptides with different hydrophobic residues induced by metal-ion binding. Biophys. Chem. 277, 106661.
Hayashi, T., Kawasaki, M., Kamatari, Y.O., and Oda, M. (2021) Single-chain Fv antibody covalently linked to antigen peptides and its structural evaluation. Anal. Biochem. 629, 114312.
Oda, M. (2021) Structural basis for Ca2+-dependent catalysis of a cutinase-like enzyme and its engineering: application to enzymatic PET depolymerization. Biophys. Physicobiol. 18, 168-176. [PDF]
Kawasaki, M. and Oda, M. (2021) DNA-binding function of c-Myb R2R3 around thermal denaturation temperature. Biophys. Physicobiol. 18, 78-84. [PDF]
Liao, Z., Senga, A., Kawai, F., and Oda, M. (2020) Effects of residue 107 of the PET hydrolase Cut190 on its activity and thermal stability. Current Topics in Peptide & Protein Research 21, 69-74.
Emori, M.*, Numoto, N.*, Senga, A., Bekker, G.-J., Kamiya, N., Kobayashi, Y., Ito, N., Kawai, F., and Oda, M. (2021) Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability. Proteins 89 (5), 502-511. (*equal contributors)
Iiyama, M.*, Numoto, N.*, Ogawa, S., Kuroda, M., Morii, H., Abe, R., Ito, N., and Oda, M. (2021) Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains. Mol. Immunol. 131, 51-59. (*equal contributors)
Senga, A.*, Numoto, N.*, Yamashita, M., Iida, A., Ito, N., Kawai, F., and Oda, M. (2021) Multiple structural states of Ca2+ regulated PET hydrolase, Cut190, and its correlation with activity and stability. J. Biochem. 169 (2) 207-214. (*equal contributors)
Kawai, F., Kawabata, T., and Oda, M. (2020) Current state and perspectives related to the PET hydrolases available for biorecycling. ACS Sustainable Chem. Eng. 8 (24), 8894-8908.
Kawasaki, M., Hosoe, Y., Kamatari, Y.O., and Oda, M. (2020) Naive balance between structural stability and DNA-binding ability of c-Myb R2R3 under physiological ionic conditions. Biophys. Chem. 258, 106319.
Yamaoka, T., Kamatari, Y.O., Maruno, T., Kobayashi, Y., and Oda, M. (2020) Structural and functional evaluation of single-chain Fv antibody HyC1 recognizing the residual native structure of hen egg lysozyme. Biosci. Biotechnol. Biochem. 84 (2), 358-364.
Nishiguchi, A.*, Numoto, N.*, Ito, N., Azuma, T., and Oda, M. (2019) Three-dimensional structure of a high affinity anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain. Mol. Immunol. 114, 545-552. (*equal contributors)
Senga, A.*, Hantani, Y.*, Bekker, G.-J., Kamiya, N., Kimura, Y., Kawai, F., and Oda, M. (2019) Metal binding to cutinase-like enzyme from Saccharomonospora viridis AHK190 and its effects on enzyme activity and stability. J. Biochem. 166 (2), 149-156. (*equal contributors)
Inaba, S., Shiota, S., Yoshida, T., and Oda, M. (2019) Site-specific observation of the conformational change of a protein with 15N-labeled Tyr residues using NMR. Anal. Biochem. 574, 34-38.
Hosoe, Y., Numoto, N., Inaba, S., Ogawa, S., Morii, H., Abe, R., Ito, N., and Oda, M. (2019) Structural and functional properties of Grb2 SH2 dimer in CD28 binding. Biophys. Physicobiol. 16, 80-88. [PDF]
Hantani, Y., Imamura, H., Yamamoto, T., Senga, A., Yamagami, Y., Kato, M., Kawai, F., and Oda, M. (2018) Functional characterizations of polyethylene terephthalate-degrading cutinase-like enzyme Cut190 mutants using bis(2-hydroxyethyl) terephthalate as the model substrate. AIMS Biophysics 5 (4), 290-302.
Oda, M., Yamagami, Y., Inaba, S., Oida, I., Yamamoto, M., Kitajima, S., and Kawai, F. (2018) Enzymatic hydrolysis of PET: Functional roles of three Ca2+ ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity. Appl. Microbiol. Biotech. 102 (23), 10067-10077.
Numoto, N., Kamiya, N., Bekker, G.-J., Yamagami, Y., Inaba, S., Ishii, K., Uchiyama, S., Kawai, F., Ito, N., and Oda, M. (2018) Structural dynamics of the PET-degrading cutinase- like enzyme from Saccharomonospora viridis AHK190 in substrate-bound states elucidates the Ca2+-driven catalytic cycle. Biochemistry 57 (36), 5289-5300.
Hantani, Y., Motoki, S., Miyagawa, A, Yamamura, H., and Oda, M. (2018) Transglycosylation activity of catalytic domain mutant of endo-1,3-β-glucanase from Cellulosimicrobium cellulans. Protein & Peptide Letters 25 (8), 734-739.
Hosoe, Y.*, Inaba, S.*, Sekiguchi, H., Sasaki, Y.C., and Oda, M. (2018) DNA-binding induced conformational change of c-Myb R2R3 analyzed using diffracted X-ray tracking. Biochem. Biophys. Res. Commun. 503, 338-343. (*equal contributors)
Shiota, A., Inaba, S., and Oda, M. (2018) Effects of active site residues of 3α-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831 on its catalysis and cofactor binding. Biosci. Biotechnol. Biochem. 82 (10), 1702-1707.
Inaba, S., Kamiya, N., Bekker, G.-J., Kawai, F., and Oda, M. (2019) Folding thermodynamics of PET-hydrolyzing enzyme Cut190 depending on Ca2+ concentration. J. Therm. Anal. Calorim. 135 (5), 2655-2663.
Oda, M., Xi, Z., Inaba, S., Slack, R.L., and Ishima, R. (2019) Binding thermodynamics of metal ions to HIV-1 ribonuclease H domain. J. Therm. Anal. Calorim. 135 (5), 2647-2653.
Oda, M., Inaba, S., Kamiya, N., Bekker, G.-J., and Mikami, B. (2018) Structural and thermodynamic characterization of endo-1,3-β-glucanase: Insights into the substrate recognition mechanism. BBA – Proteins and Proteomics 1866 (3), 415-425.
Inaba, S., Fukada, H., and Oda, M. (2018) Effect of a salt-bridge between inter-repeats on the 3D structure of the c-Myb DNA-binding domain revealed by thermodynamic analysis. J. Therm. Anal. Calorim. 131 (1), 335-341.
Usui, D., Inaba, S., Sekiguchi, H., Sasaki, Y. C., Tanaka, T., and Oda, M. (2017) First observation of metal ion-induced structural fluctuations of α-helical peptides by using diffracted X-ray tracking. Biophys. Chem. 228, 81-86.
Usui, D., Inaba, S., Kamatari, Y. O., Ishiguro, N., and Oda, M. (2017) Light-chain residue 95 is critical for antigen binding and multispecificity of monoclonal antibody G2. Biochem. Biophys. Res. Commun. 490, 1205-1209.
Sato, Y., Inaba, S., Fukada, H., Azuma, T., and Oda, M. (2017) Pronounced effect of hapten binding on thermal stability of an anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain. Mol. Immunol. 85, 130-136.
Kawabata, T., Oda, M., and Kawai, F. (2017) Mutational analysis of cutinase-like enzyme, Cut190, based on the 3D docking structure with model compounds of polyethylene terephthalate. J. Biosci. Bioeng. 124 (1), 28-35.
Miki, A., Inaba, S., Maruno, T., Kobayashi, Y., and Oda, M. (2017) Tryptophan introduction can change β-glucan binding ability of the carbohydrate-binding module of endo-1,3-β-glucanase. Biosci. Biotechnol. Biochem. 81 (5), 951-957.
Inaba, S.*, Numoto, N.*, Ogawa, S., Morii, H., Ikura, T., Abe, R., Ito, N., and Oda, M. (2017) Crystal structures and thermodynamic analysis reveal distinct mechanisms of CD28 phosphopeptide binding to the Src homology 2 (SH2) domains of three adaptor proteins. J. Biol. Chem. 292 (3), 1052-1060. (*equal contributors)
Sato, Y.*, Tanaka, Y.*, Inaba, S., Sekiguchi, H., Maruno, T., Sasaki, Y.C., Fukada, H., Kobayashi, Y., Azuma, T., and Oda, M. (2016) Structural dynamics of a single-chain Fv antibody against (4-hydroxy-3-nitrophenyl)acetyl. Int. J. Biol. Macromol. 91, 151-157. (*equal contributors)
Oda, M. and Kuroda, M. (2016) Molecular dynamics simulations of inclusion complexation of glycyrrhizic acid and cyclodextrins (1:1) in water. J. Incl. Phenom. Macro. Chem. 85, 271-279.
Oda, M., Tsumuraya, T, and Fujii, I. (2016) Effects of substrate conformational strain on binding kinetics of catalytic antibody. Biophys. Physicobiol. 13, 135-138. [PDF]
Oda, M., and Azuma, T. (2016) Affinity maturation of anti-(4-hydroxy-3-nitrophenyl)acetyl antibodies accompanies a modulation of antigen specificity. Mol. Immunol. 270, 8-12.
Inaba, S., Fukada, H., and Oda, M. (2016) Folding thermodynamics of c-Myb DNA-binding domain in correlation with its α-helical contents. Int. J. Biol. Macromol. 82, 725-732.
Inaba, S., Maeno, A., Sakurai, K., Puthenpurackal, S. N., Ikegami, T., Akasaka, K., and Oda, M. (2015) Functional conformer of c-Myb DNA-binding domain revealed by variable temperature studies. FEBS J. 282 (23), 4497-4514.[PDF]
Inaba, S., Fukada, H., and Oda, M. (2016) Thermodynamic effects of a linker region between two repeats of a protein, c-Myb R2R3, on its stability and structural dynamics. J. Therm. Anal. Calorim. 123, 1763-1767.
Miki, A.*, Inaba, S.*, Baba, T., Kihira, K., Fukada, H., and Oda, M. (2015) Structural and physical properties of collagen extracted from moon jellyfish under neutral pH conditions. Biosci. Biotechnol. Biochem. 79 (10), 1603-1607. (*equal contributors)
Adhikary, R., Yu, W., Oda, M., Walker, R.C., Chen, T., Stanfield, R.L., Wilson, I.A., Zimmermann, J., and Romesberg, F.E. (2015) Adaptive mutations alter antibody structure and dynamics during affinity maturation. Biochemistry 54 (11), 2085-2093.
Kawai, F., Oda, M., Tamashiro, T., Waku, T., Tanaka, N., Yamamoto, M., Mizushima, H., Miyakawa, T., and Tanokura, M. (2014) A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotech. 98 (24), 10053-10064.
Kamatari, Y. O., Ohta, S., Inoshima, Y., Oda, M., Maruno, T., Kobayashi, Y., and Ishiguro, N. (2014) Identification and characterization of a multispecific monoclonal antibody G2 directed against chicken prion protein. Protein Sci. 23 (8), 1050-1059.
Izutani, Y. Kanaori, K., and Oda, M. (2014) Aggregation property of glycyrrhizic acid and its interaction with cyclodextrins analyzed by dynamic light scattering, isothermal titration calorimetry, and NMR. Carbohydr. Res. 392, 25-30.
Inaba, S., Fukada, H., Ikegami, T., and Oda, M. (2013) Thermodynamic effects of multiple protein conformations on stability and DNA binding. Arch. Biochem. Biophys. 537 (2), 225-232.
Adhikary, R., Yu, W., Oda, M., Zimmermann, J., and Romesberg, F.E. (2012) Protein dynamics and the diversity of an antibody response. J. Biol. Chem. 287 (32), 27139-27147. [PDF]
Tamashiro, T., Tanabe, Y., Ikura, T., Ito, N., and Oda, M. (2012) Critical roles of Asp270 and Trp273 in the α-repeat of the carbohydrate-binding module of endo-1,3-β-glucanase for laminarin-binding avidity. Glycoconj. J. 29 (1), 77-85.
Tanabe, Y., and Oda, M. (2011) Molecular characterization of endo-1,3-β-glucanase from Cellulosimicrobium cellulans: effects of carbohydrate-binding module on enzymatic function and stability. Biochimica et Biophysica Acta – Proteins and Proteomics, 1814 (12), 1713-1719.
Oda, M., Kitai, A., Murakami, A., Nishimura, M., Ohkuri, T., Abe, Y., Ueda, T., Nakamura, H., and Azuma, T. (2010) Evaluation of the conformational equilibrium of reduced hen egg lysozyme by antibodies to the native form. Arch. Biochem. Biophys. 494 (2), 145-150.
Oda, M., Saito, M., Tsumuraya, T, and Fujii, I. (2010) Contribution of the trifluoroacetyl group in the thermodynamics of antigen-antibody binding. J. Mol. Recognit. 23 (3), 263-270.
Oda, M.*, Uchiyama, S.*, Noda, M., Nishi, Y., Koga, M., Mayanagi, K., Robinson, C.V., Fukui, K., Kobayashi, Y., Morikawa, K., and Azuma, T. (2009) Effects of antibody affinity and antigen valence on the molecular forms of immune complexes. Mol. Immunol. 47 (2-3), 357-364. (*equal contributors)
Tanabe, Y., Pang, Z., and Oda, M. (2008) Cloning and sequencing of endo-1,3-β-glucanase from Cellulosimicrobium cellulans. J. Biol. Macromol. 8 (3), 60-63. [PDF]
Thielges, M.C., Zimmermann, J., Yu, W., Oda, M., and Romesberg, F.E. (2008) Exploring the energy landscape of antibody-antigen complexes: Protein dynamics, flexibility, and molecular recognition. Biochemistry 47 (27), 7237-7247.
Oda, M., Ito, N., Tsumuraya, T., Suzuki, K., Sakakura, M., and Fujii, I. (2007) Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis. J. Mol. Biol. 369 (1), 198-209.
Nakamura, S., Oda, M., Kataoka, S., Ueda, S., Uchiyama, S., Yoshida, T., Kobayashi, Y., and Ohkubo, T. (2006) Apo- and holo-structures of 3α-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831: Loop-helix transition induced by coenzyme binding. J. Biol. Chem. 281 (42), 31876-31884. [PDF]
Kataoka, S., Nakamura, S., Ohkubo, T., Ueda, S., Uchiyama, S., Kobayashi, Y., and Oda, M. (2006) Crystallization and preliminary X-ray analysis of the complex of NADH and 3α-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831. Acta Crystallogr. F62, 569-571. [PDF]
Oda, M., Uchiyama, S., Robinson, C.V., Fukui, K., Kobayashi, Y., and Azuma, T. (2006) Regional and segmental flexibility of antibodies in interaction with antigens of different size. FEBS J. 273 (7), 1476-1487.
Izutani, Y., Kanaori, K., Imoto, T., and Oda, M. (2005) Interaction of gymnemic acid with cyclodextrins analyzed by isothermal titration calorimetry, NMR, and dynamic light scattering. FEBS J. 272 (23), 6154-6160. [PDF]
Pang, Z., Otaka, K., Maoka, T., Hidaka, K., Ishijima, S., Oda, M., and Ohnishi, M. (2005) Structure of β-glucan oligomer from laminarin and its effect on human monocytes to inhibit the proliferation of U937 cells. Biosci. Biotechnol. Biochem. 69 (3), 553-558. [PDF]
Saito, M., Okazaki, I., Oda, M., and Fujii, I. (2005) A free energy calculation study of the effect of H→F substitution on binding affinity in ligand-antibody interactions. J. Comp. Chem. 26 (3), 272-282.
Pang, Z., Kang, Y.-N., Ban, M., Oda, M., Kobayashi, R., Ohnishi, M., and Mikami, B. (2005) Crystallization and preliminary crystallographic analysis of endo-1,3-β-glucanase from Arthrobacter sp. Acta Crystallog. F61, 68-70. [PDF]
Sagawa, T., Oda, M., Morii, H., Takizawa, H., Kozono, H., and Azuma, T. (2005) Conformational changes in the antibody constant domains upon hapten binding. Mol. Immunol. 42 (1), 9-18.
Saito, K.*, Oda, M.*, Sarai, A., Azuma, T, and Kozono, H. (2004) Bound peptide-dependent thermal stability of major histocompatibility complex class II molecule I-Ek. Biochemistry 43 (31), 10186-10191. (*equal contributors)
Oda, M., Sato-Nakamura, N., and Azuma, T. (2004) Molecular characterization of monovalent and multivalent hapten-protein conjugates for analysis of the antigen-antibody interaction. Anal. Biochem. 333 (2), 365-371.
Ueda, S., Oda, M., Imamura, S., and Ohnishi, M. (2004) Kinetic study of the enzymatic cycling reaction conducted with 3α-hydroxysteroid dehydrogenase in the presence of excessive thio-NAD+ and NADH. Anal. Biochem. 332 (1), 84-89.
Ueda, S., Oda, M., Imamura, S., and Ohnishi, M. (2004) Transient-phase kinetic studies on the nucleotide binding to 3α-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831 using fluorescence stopped-flow procedures. Eur. J. Biochem. 271 (9), 1774-1780. [PDF]
Saito, K., Oda, M., Sarai, A., Azuma, T, and Kozono, H. (2004) Contribution of a single hydrogen bond between βHis81 of MHC class II I-Ek and the bound peptide to the pH-dependent thermal stability. Micro. Immunol. 48 (1), 53-57.
Tobita, T.*, Oda, M.*, and Azuma, T. (2004) Segmental flexibility and avidity of IgM in the interaction of polyvalent antigens. Mol. Immunol. 40 (11), 803-811. (*equal contributors)
Saito, K., Sarai, A., Oda, M., Azuma, T, and Kozono, H. (2003) Thermodynamic analysis of the increased stability of major histocompatibility complex class II molecule I-Ek complexed with an antigenic peptide at acidic pH. J. Biol. Chem. 278 (17), 14732-14738. [PDF]
Oda, M., Kozono, H., Morii, H., and Azuma, T. (2003) Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int. Immunol. 15 (3), 417-426. [PDF]
Sagawa, T., Oda, M., Ishimura, M., Furukawa, K., and Azuma, T. (2003) Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten. Mol. Immunol. 39 (13), 801-808.
Tobita, T., Oda, M., Morii, H., Kuroda, M., Yoshino, A., Azuma, T., and Kozono, H. (2003) A role for the P1 anchor residue in the thermal stability of MHC class II molecule I-Ab. Immunol. Lett. 85 (1), 47-52.
Oda, M., and Azuma, T. (2000) Reevaluation of stoichiometry and affinity/avidity in interactions between anti-hapten antibodies and mono- or multi-valent antigens. Mol. Immunol. 37 (18), 1111-1122.
Oda, M., and Nakamura, H. (2000) Thermodynamic and kinetic analyses for understanding sequence-specific DNA recognition. Genes to Cells 5 (5), 319-326.
Oda, M., Furukawa, K., Sarai, A., and Nakamura, H. (1999) Construction of an artificial tandem protein of the c-Myb DNA-binding domain and analysis of its DNA binding specificity. Biochem. Biophys. Res. Commun. 262 (1), 94-97.
Oda, M., Furukawa, K., Sarai, A., and Nakamura, H. (1999) Kinetic analysis of DNA binding by the c-Myb DNA-binding domain using surface plasmon resonance. FEBS Lett. 454 (3), 288-292.
Oda, M., Shiraishi, A., and Hasegawa, M. (1998) Analysis of the ternary complex formation of human urokinase with the separated two domains of its receptor. Eur. J. Biochem. 256 (2), 411-418. [PDF]
Oda, M., Furukawa, K., Ogata, K., Sarai, A., and Nakamura, H. (1998) Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 276 (3), 571-590.
Oda, M., Furukawa, K., Ogata, K., Sarai, A., Ishii, S., Nishimura, Y., and Nakamura, H. (1997) Identification of indispensable residues for specific DNA-binding in the imperfect tandem repeats of c-Myb R2R3. Protein Eng. 10 (12), 1407-1414.(Cover Illustration) [PDF]
Oda, M., Furukawa, K., Ogata, K., Sarai, A., Ishii, S., Nishimura, Y., and Nakamura, H. (1997) Investigation of the pyrimidine preference by the c-Myb DNA-binding domain at the initial base of the consensus sequence. J. Biol. Chem. 272 (29), 17966-17971. [PDF]
Last updated; October 1, 2023

